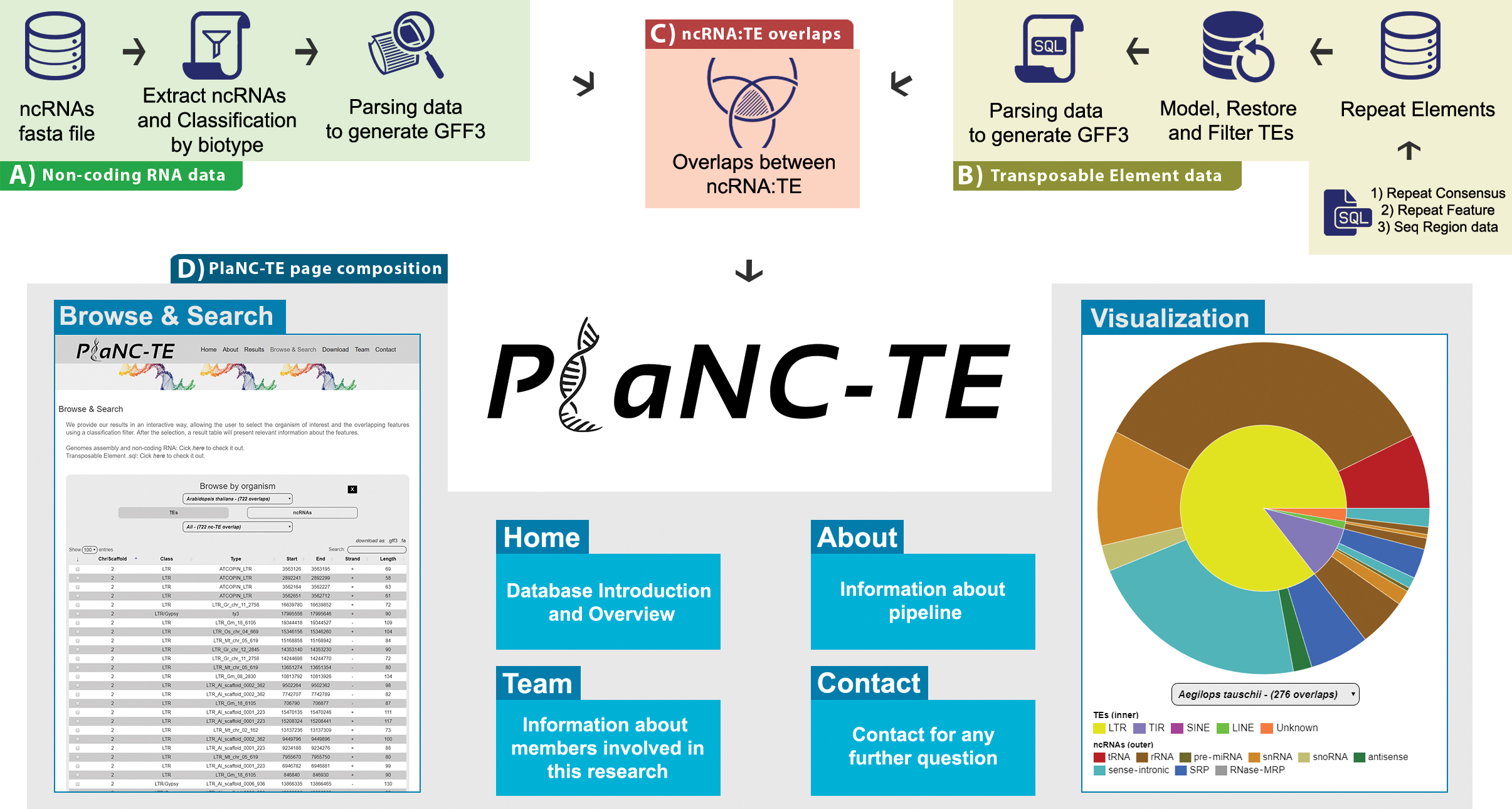
Non-coding RNAs (ncRNAs) are transcripts that do not encode proteins, with a highly diverse set of functions, acting as components of complex assemblies (e.g. rRNAs, tRNAs and snRNAs), catalytic particles (ribozymes), and guiding molecules (miRNAs, siRNAs, piRNAs and gRNAs). Transposable Elements (TEs) are ubiquitous mobile genetic elements (MGEs) in eukaryotic genomes. They comprise 45% of the human genome, and in plants they can comprise up to 90%. For a long time TEs were not recognized as relevant components of their host genomes. On the contrary, they were even considered “junk DNA”. With increasing evidence supporting the important role of TEs as “drivers” of genome evolution, the scientific community has recently intensified its interest on the topic. Some studies have shown, for example, that these genomic features are significant reservoirs of non-coding features with regulatory functions (miRNAs, siRNAs and piRNAs). Following these discoveries, our research group developed PlanTE-MIR DB, the first public database that presents relationship data between miRNA and TEs in plants.
We now expand the knowledge about the relationship between TEs and ncRNAs, creating an improved successor for PlanTE-MIR DB: the PlaNC-TE DB. This portal delivers the overlapping regions between TE features and all classes of ncRNAs available at Ensembl Plants in an user friendly way, allowing the users to download bulk information for each genome and to interactively search for results.